Using VTK files in Paraview#
Loading Correlation Data#
The image correlation scripts spam-ldic, spam-ddic and spam-gdic all can produce VTK files. These files can be loaded into Paraview (Currently at version 5.6.0) for easy visalisation. This is super convenient, but a number of things need to be known:
Paraview is good at rendering meshed objects, and so there is a clear distinction between quantities that are stored at the nodes of the mesh, and quantities that are stored in the elements of the mesh.
In spam-ldic it is not obvious to us what the best way to save the measured displacements (it depends on whether you think of each measurement as representing a subvolume or a point), we have elected to store the results as elements or “cells”.
For the other two techniques the displacements are stored at points.
To make nice visualisations of vector fields, drag and drop your VTKs into Paraview (if you drop multiple with similar names they will be opened as a time series).
press on the calculator and make a field called “mag(displacements)” - cells for ldic and points for the others - whose value is “mag(displacements)”
Now press on the glyph button - looks like a sphere with points - it’s on “arrows” by default
Orientation array = displacements
Scale array = mag(displacements)
Scale Factor = However much you want to exaggerate your vectors by. 1 → real size wrt. image size
Glyph Mode = “All Points”
Coloring = mag(displacements)
Now apply a threshold on the return status = minimum 2 to exclude all non-correlating points (not needed in global of course)
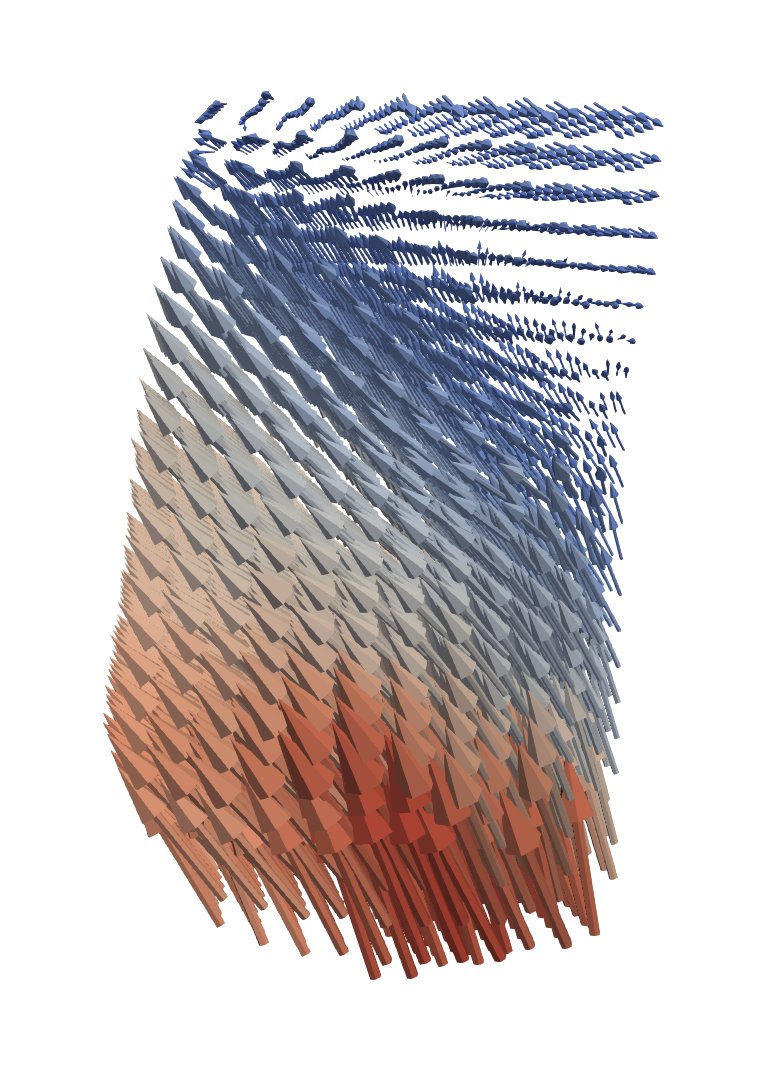
Displacement arrows obtained with spam-ldic#

Displacement arrows obtained with spam-ddic on same data#
Comparing to your original image#
Since the kinematic fields shown above are expressed in the original image coordinates, Paraview offers a good place to plot the original tomography data together with this measurement.
Danger
If you just drag-and-drop your TIFF file into Paraview bad things happen:
The y-direction is flipped, so all comparison with kinematic fields is meaningless
The imported data is considered as point-wise data and is consequently interpolated if you slice it
To avoid these two problems, the TIFF file can be converted into cell data and resaved:
import tifffile
import spam.helpers
greys = tifffile.imread("M2EA05-01.tif")
spam.helpers.writeStructuredVTK(cellData={'grey':greys}, fileName="M2EA05-01.vtk")
This makes a rather huge and redundant VTK, but it’s worth it. There is also an external script which uses another approach but has the same result: spam.helpers.vtkio.TIFFtoVTK.

Displacement arrows obtained with spam-ddic with original greyscale (rendered with threshold)#
