Note
Go to the end to download the full example code.
Kalisphera virtual concrete generation for multimodal registration#
In this example we will read a series of sphere centres and radii coming from a DEM simulation and generate two voxelised images:
The first will correspond to a neutron concrete image
The second will correspond to an x-ray concrete image.
For both images, we will use Kalisphera to make a “perfect” rendering, which we will then degrade with blur and noise to make it look like a “real” tomography image. The technique is detailed in [Tengattini2015].
Note
The two images will not be aligned. The spheres of the neutron concrete image will be displaced to generate the x-ray concrete image. This is a very convenient starting point to then do a multimodal registration of the two images.
import numpy
import scipy.ndimage
import spam.datasets
import spam.deformation
import spam.mesh
import spam.kalisphera
import matplotlib.pyplot as plt
#import tifffile
#import spam.helpers
Create a helper function#
First, we will need a helper function that creates the cylindrical virtual “concrete”:
def virtualConcrete(dims, phaseColours, positions1, radii1, positions2, radii2, cylinderRadius, cylinderCentre, cylinderHeight, gaussianBlur=0.8, gaussianNoise=0.2):
"""
This function makes a z-aligned cylindrical virtual "concrete" with a matrix and embedded spheres
of different colours. Background colour = 0.0.
``Kalisphera`` is used to make a "perfect" rendering of the spheres, which will then be degraded with blur and noise to make it look like a “real” tomography image
Parameters:
-----------
dims : 3-component list of output dimensions
phaseColours: 3-component list
values for 3 phases: matrix, grains and pores
positions1 : Nx3 numpy array
positions of grain phase spheres
radii1 : N numpy array
radii of grain phase spheres
positions2 : Nx3 numpy array
positions of pores phase spheres
radii2 : N numpy array
radii of pores phase spheres
cylinderRadius : float
cylinderCentre : 3-component list
cylinderHeight : float
Returns:
--------
imCyl : 3D numpy array
"""
phaseOne = numpy.zeros(dims, dtype="<f8")
phaseTwo = numpy.zeros(dims, dtype="<f8")
# TODO top and bottom crop
imCyl = (spam.mesh.createCylindricalMask(dims, cylinderRadius, centre=cylinderCentre[1:]) * phaseColours[0]).astype('<f8')
spam.kalisphera.makeSphere(phaseOne, positions1, radii1)
spam.kalisphera.makeSphere(phaseTwo, positions2, radii2)
# cleanup outside of balls
phaseOne[imCyl==0] = 0
phaseTwo[imCyl==0] = 0
# Add in contributions
imCyl += phaseOne * (phaseColours[1] - phaseColours[0])
imCyl += phaseTwo * (phaseColours[2] - phaseColours[0])
# brutally and horizontally slice the height limits
imCyl[0:int(cylinderCentre[0]-cylinderHeight/2)] = 0
imCyl[ int(cylinderCentre[0]+cylinderHeight/2)::] = 0
# Add gaussian (spatial) blur
imCyl = scipy.ndimage.gaussian_filter(imCyl, sigma=gaussianBlur)
# Add random noise
imCyl = numpy.random.normal(imCyl, scale=gaussianNoise)
return imCyl
Assign phase values#
We assume that concrete is a three-phase material: mortar, grains, pores. To create the neutron and the x-ray concrete images, we will need to assign reasonable values for each phase imaged with the two modalities:
# Reasonable values for each phase of concrete
mortarX = 3 # X-ray , mortar
mortarN = 5 # Neutron, mortar
grainsX = 5.2 # X-ray , grains
grainsN = 0.2 # Neutron, grains
poresX = 0.1 # X-ray , pores
poresN = 0.1 # Neutron, pores
We will then set the blur and noise levels to make these values look like a “real” tomography image
# The standard deviation of the image blurring to be applied first
blurSTD = 0.5
# The standard deviation of the random noise to be added to each voxel
noiseSTD = 0.05
Read input DEM data#
We will now read the spheres positions and radii coming from a DEM simulation. These data will be the input for the neutron image:
spheres = spam.datasets.loadDEMspheresMMR()
pos = spheres[:, -2::-1] # swap z and x axis
radii = spheres[:, -1]
The DEM data is normally in SI units, so we need to define the pixel size for voxelisation:
pixelSizeM = 0.00005
posPx = pos/pixelSizeM
radiiPx = radii/pixelSizeM
We will set the dimensions of our cylinders and center the spheres to stick in the middle:
dims = (numpy.ceil(numpy.max(posPx, axis=0) - numpy.min(posPx, axis=0))*1.3).astype(int)
middle = (dims - 1) / 2.0
cylRadiusPx = min(dims[1:])*0.9/2.0
# Center the spheres and stick in middle of dims
posPx -= numpy.mean(posPx, axis=0)
posPx += middle
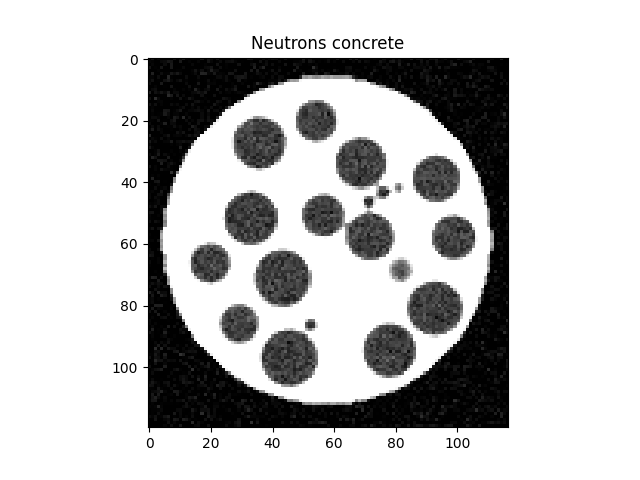
Create neutron image#
We will now call the helper function to create the neutron image:
# Create the neutrons
neutrons = virtualConcrete(dims, [mortarN, grainsN, poresN],
posPx[radiiPx==radiiPx.max()], radiiPx[radiiPx==radiiPx.max()],
posPx[radiiPx==radiiPx.min()], radiiPx[radiiPx==radiiPx.min()],
cylRadiusPx, middle, dims[0]*0.9,
gaussianBlur=blurSTD, gaussianNoise=noiseSTD)
#tifffile.imwrite("Neutrons.tif", neutrons.astype('<f4'))
plt.figure()
plt.imshow(neutrons[neutrons.shape[0] // 2], vmin=0, vmax=1, cmap='Greys_r')
plt.title("Neutrons concrete")
plt.show()
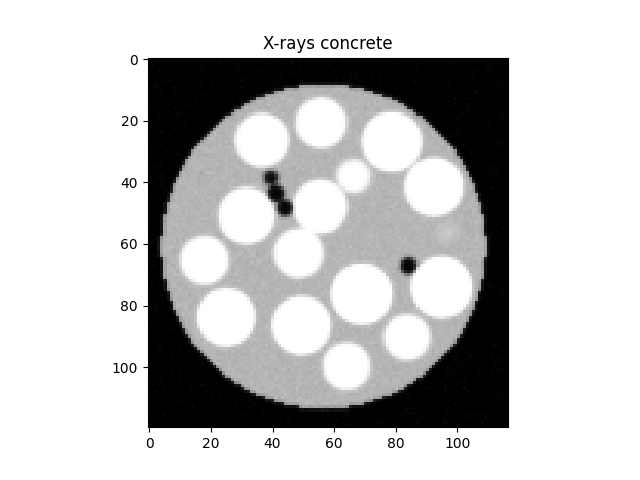
Create x-ray image#
We will now set the positions of the spheres for the x-ray image. For this, we will displace the spheres positions coming from the DEM simulation based on a deformation function that corresponds to a rigid body motion:
# move the grains according to the deformation function
Phi = spam.deformation.computePhi({'t': [0, 2, -1], 'r': [90, 0, 0]})
for i, poPx in enumerate(posPx):
posPx[i] += spam.deformation.decomposePhi(Phi, PhiCentre=middle, PhiPoint=poPx)['t']
# moving the middle of the cylinder
middleMoved = middle + spam.deformation.decomposePhi(Phi)['t']
We will now call the helper function to create the x-rays image:
# Create the x-rays
xrays = virtualConcrete(dims, [mortarX, grainsX, poresX],
posPx[radiiPx==radiiPx.max()], radiiPx[radiiPx==radiiPx.max()],
posPx[radiiPx==radiiPx.min()], radiiPx[radiiPx==radiiPx.min()],
cylRadiusPx, middleMoved, dims[0]*0.9,
gaussianBlur=blurSTD, gaussianNoise=noiseSTD)
#tifffile.imwrite("Xrays.tif", xrays.astype('<f4'))
plt.figure()
plt.imshow(xrays[xrays.shape[0] // 2], vmin=0, vmax=5, cmap='Greys_r')
plt.title("X-rays concrete")
plt.show()
The datasets are ready!
Initial guess for multimodal registration#
These two images can now be the input of a mulitmodal registration algorithm. To help this algorithm to converge, we can give a reasonable initial guess of the transformaion between these two images:
# PhiGuess: a bit off of the actual Phi that displaced the spheres
PhiGuess = spam.deformation.computePhi({'t': [0, 1, 0], 'r': [91, 0, 0]})
We can now save this initial guess in a TSV file, to be a ready input for the mulitomodal registration algorithm:
# Creating a dictionary emulating a converged previous registration.
Reg = {}
Reg['Phi'] = PhiGuess
Reg['error'] = 0.001
Reg['returnStatus'] = 2
Reg['iterations'] = 2
Reg['deltaPhiNorm'] = 0.01
#spam.helpers.writeRegistrationTSV('./guessPhiNtoX.tsv', middle, Reg)
Total running time of the script: (0 minutes 0.819 seconds)
